[ET Trac] #2599: nsnstohmns cannot be reproduced using LORENE2
Roland Haas
trac-noreply at einsteintoolkit.org
Thu Mar 10 14:09:09 CST 2022
#2599: nsnstohmns cannot be reproduced using LORENE2
Reporter: Ken Hui
Status: open
Milestone: ET_2021_05
Version: ET_2021_05
Type: bug
Priority: minor
Component: Cactus
Comment (by Roland Haas):
Thanks for the update. The changes in solar mass and and gravitational constant I can understand \(and expect\). Though how can MeV in Joule change? It’s essentially the electron charge which I would have expected to be very well known.
I agree that the plots are not directly useful. The interesting bit would be that failure happens once the central density rises ie star start to interact.
Another useful plot, since the grid structure is what fails would be a plot of the grid structure. Eg using the plot-bboxes tool in [https://bitbucket.org/eschnett/carpet/src/master/Carpet/src/util/plot-bboxes](https://bitbucket.org/eschnett/carpet/src/master/Carpet/src/util/plot-bboxes) It expects as input one of Carpet’s `carpet-grid-structure` files:
```
> gnuplot
$ load "plot-bboxes"
```
then paste \(since it reads from `/dev/tty`\) one of the sections from the grid structure file, eg:
```
iteration 0
maps 1
0 mglevels 1
0 0 reflevels 3
0 0 0 components 2
0 0 0 0 0 ([0,0,0]:[136,136,68]:[4,4,4]/[0,0,0]:[34,34,17]/[35,35,18]/22050) [[1,1,1],[1,1,0]]
0 0 0 1 1 ([0,0,72]:[136,136,136]:[4,4,4]/[0,0,18]:[34,34,34]/[35,35,17]/20825) [[1,1,0],[1,1,1]]
0 0 1 components 2
0 0 1 0 0 ([36,36,36]:[100,100,68]:[2,2,2]/[18,18,18]:[50,50,34]/[33,33,17]/18513) [[0,0,0],[0,0,0]]
0 0 1 1 1 ([36,36,70]:[100,100,100]:[2,2,2]/[18,18,35]:[50,50,50]/[33,33,16]/17424) [[0,0,0],[0,0,0]]
0 0 2 components 2
0 0 2 0 0 ([52,52,52]:[84,84,68]:[1,1,1]/[52,52,52]:[84,84,68]/[33,33,17]/18513) [[0,0,0],[0,0,0]]
0 0 2 1 1 ([52,52,69]:[84,84,84]:[1,1,1]/[52,52,69]:[84,84,84]/[33,33,16]/17424) [[0,0,0],[0,0,0]]
```
which should produce this plot:
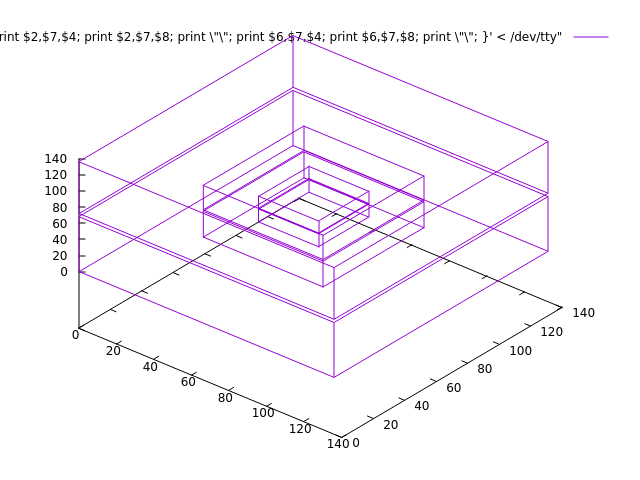
This may also be possible with a dedicated Cactus analysis package like SimulationTools or Kuibit or PyCactus/PostCactus listed on [https://docs.einsteintoolkit.org/et-docs/Analysis\_and\_post-processing](https://docs.einsteintoolkit.org/et-docs/Analysis_and_post-processing) .
--
Ticket URL: https://bitbucket.org/einsteintoolkit/tickets/issues/2599/nsnstohmns-cannot-be-reproduced-using
-------------- next part --------------
An HTML attachment was scrubbed...
URL: http://lists.einsteintoolkit.org/pipermail/trac/attachments/20220310/d4f10f6c/attachment.html
More information about the Trac
mailing list